Combining DNA Fragments With today's technologies, scientists can produce custom-built DNA molecules in the lab and then insert those molecules—along with the genes they carry—into living cells. The first step in this sort of genetic engineering is to build a DNA sequence with the gene or genes you'd like to insert into a cell. Machines known as DNA synthesizers can produce short pieces of DNA, up to several hundred bases in length. These synthetic sequences can then be joined to natural sequences using DNA ligase or other enzymes that splice DNA together. These same enzymes make it possible to take a gene from one organism and attach it to the DNA of another organism, as shown in Figure 15–8. The resulting molecules are called recombinant DNA. This technology relies on the fact that any pair of complementary sequences tends to bond, even if each sequence comes from a different organism.  Recombinant-DNA technology—joining together DNA from two or more sources—makes it possible to change the genetic composition of living organisms. By manipulating DNA in this way, scientists can investigate the structure and functions of genes.
Recombinant-DNA technology—joining together DNA from two or more sources—makes it possible to change the genetic composition of living organisms. By manipulating DNA in this way, scientists can investigate the structure and functions of genes.
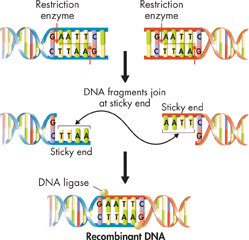
FIGURE 15–8 Joining DNA Pieces Together Recombinant DNA molecules are made up of DNA from different sources. Restriction enzymes cut DNA at specific sequences, producing “sticky ends,” which are single-stranded overhangs of DNA. If two DNA molecules are cut with the same restriction enzyme, their sticky ends will bond to a fragment of DNA that has the complementary sequence of bases. An enzyme known as DNA ligase can then be used to join the two fragments.
dPlasmids and Genetic Markers Scientists working with recombinant DNA soon discovered that many of the DNA molecules they tried to insert into host cells simply vanished because the cells often did not copy, or replicate, the added DNA. Today scientists join recombinant DNA to another piece of DNA containing a replication “start” signal. This way, whenever the cell copies its own DNA, it copies the recombinant DNA too.
In addition to their own chromosomes, some bacteria contain small circular DNA molecules known as plasmids. Plasmids, like those shown in Figure 15–9, are widely used in recombinant DNA studies. Joining DNA to a plasmid, and then using the recombinant plasmid to transform bacteria, results in the replication of the newly added DNA along with the rest of the cell's genome.
Plasmids are also found in yeasts, which are single-celled eukaryotes that can be transformed with recombinant DNA as well. Biologists working with yeasts can construct artificial chromosomes containing centromeres, telomeres, and replication start sites. These artificial chromosomes greatly simplify the process of introducing recombinant DNA into the yeast genome.
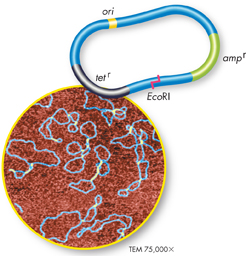
FIGURE 15–9 A Plasmid Map Plasmids used for genetic engineering typically contain a replication start signal, called the origin of replication (ori), and a restriction enzyme cutting site, such as EcoRI. They also contain genetic markers, like the antibiotic resistance genes tetr and ampr shown here.
Table of Contents
- Formulas and Equations
- Applying Formulas and Equations
- Mean, Median, and Mode
- Estimation
- Using Measurements in Calculations
- Effects of Measurement Errors
- Accuracy
- Precision
- Comparing Accuracy and Precision
- Significant Figures
- Calculating With Significant Figures
- Scientific Notation
- Calculating With Scientific Notation
- Dimensional Analysis
- Applying Dimensional Analysis