The Human Genome Project
 What were the goals of the Human Genome Project, and what have we learned so far?
What were the goals of the Human Genome Project, and what have we learned so far?
In 1990, the United States, along with several other countries, launched the Human Genome Project.  The Human Genome Project was a 13-year, international effort with the main goals of sequencing all 3 billion base pairs of human DNA and identifying all human genes. Other important goals included sequencing the genomes of model organisms to interpret human DNA, developing technology to support the research, exploring gene functions, studying human variation, and training future scientists.
The Human Genome Project was a 13-year, international effort with the main goals of sequencing all 3 billion base pairs of human DNA and identifying all human genes. Other important goals included sequencing the genomes of model organisms to interpret human DNA, developing technology to support the research, exploring gene functions, studying human variation, and training future scientists.
DNA sequencing was at the center of the Human Genome Project. However, the basic sequencing method you saw earlier can analyze only a few hundred nucleotides at a time. How, then, can the huge amount of DNA in the human genome be sequenced quickly? First, researchers must break up the entire genome into manageable pieces. By determining the base sequences in widely separated regions of a DNA strand, they can use the regions as markers, not unlike the mile markers along a road that is thousands of miles long. The markers make it possible for researchers to locate and return to specific locations in the DNA.
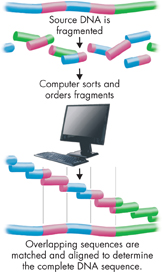
FIGURE 14–11 Shotgun Sequencing This method rapidly sorts DNA fragments by overlapping base sequences.
d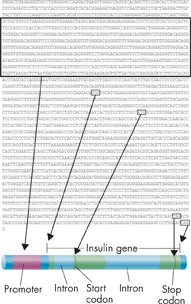
FIGURE 14–12 Locating a Gene A typical gene, such as that for insulin, has several DNA sequences that can serve as locators. These include the promoter, sequences between introns and exons, and start and stop codons.
ddSequencing and Identifying Genes Once researchers have marked the DNA strands, they can use the technique of “shotgun sequencing.” This rapid sequencing method involves cutting DNA into random fragments, then determining the base sequence in each fragment. Computer programs take the sequencing data, find areas of overlap between fragments, and put the fragments together by linking the overlapping areas. The computers then align these fragments relative to the known markers on each chromosome, as shown in Figure 14–11. The entire process is like putting a jigsaw puzzle together, but instead of matching shapes, the computer matches DNA base sequences.
Reading the DNA sequence of a genome is not the same as understanding it. Much of today's research explores the vast amount of data from the Human Genome Project to look for genes and the DNA sequences that control them. By locating sequences known to be promoters—binding sites for RNA polymerase—scientists can identify many genes. Shortly after a promoter, there is usually an area called an open reading frame, which is a sequence of DNA bases that will produce an mRNA sequence. Other sites that help to identify genes are the sequences that separate introns from exons, and stop codons located at the ends of open reading frames. Figure 14–12 shows these sites on a typical gene.
Table of Contents
- Formulas and Equations
- Applying Formulas and Equations
- Mean, Median, and Mode
- Estimation
- Using Measurements in Calculations
- Effects of Measurement Errors
- Accuracy
- Precision
- Comparing Accuracy and Precision
- Significant Figures
- Calculating With Significant Figures
- Scientific Notation
- Calculating With Scientific Notation
- Dimensional Analysis
- Applying Dimensional Analysis