Quick Lab
GUIDED INQUIRY
Modeling Restriction Enzymes
1 Write a 50-base, double-stranded DNA sequence using the bases A, C, G, and T in random order. Include each sequence shown below at least once in the sequence you write.
2 Make three copies of your double-stranded sequence on three different-colored strips of paper.
3 Use the drawings below to see how the restriction enzyme EcoRI would cut your DNA sequence. Use scissors to cut one copy of the sequence as EcoRI would.
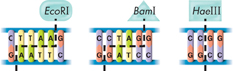
4 Use the procedure in Step 3 to cut apart another copy of your sequence as the restriction enzyme BamI would. Then, cut the third copy as the restriction enzyme HaeIII would.
5 Tape the single-stranded end of one of your DNA fragments to a complementary, single-stranded end of a classmate's fragment. This will form a single, double-stranded DNA molecule.
Observe Which restriction enzyme produced the most pieces? The fewest pieces?
Evaluate How well did your model represent the actual process of using restriction enzymes to cut DNA? (Hint: Contrast the length of your model DNA sequence to the actual length of a DNA molecule.)
Analyze and Conclude
Table of Contents
- Formulas and Equations
- Applying Formulas and Equations
- Mean, Median, and Mode
- Estimation
- Using Measurements in Calculations
- Effects of Measurement Errors
- Accuracy
- Precision
- Comparing Accuracy and Precision
- Significant Figures
- Calculating With Significant Figures
- Scientific Notation
- Calculating With Scientific Notation
- Dimensional Analysis
- Applying Dimensional Analysis